Preparations
Overview
Teaching: 45 min
Exercises: 15 minQuestions
How do I read data into R?
How do I assign variables?
What is a data frame?
How do I calculate simple statistics like mean and median?
Where can I get help?
How can I plot my data?
Objectives
Read tabular data from a file into a program.
Perform operations on a data frame of data.
Display simple graphs.
We are studying inflammation in patients who have been given a new treatment for arthritis, and need to analyze the first dozen data sets. The data sets are stored in comma-separated values (CSV) format. Each row holds the observations for just one patient. Each column holds the inflammation measured in a day, so we have a set of values in successive days. The first few rows of our first file look like this:
0,0,1,3,1,2,4,7,8,3,3,3,10,5,7,4,7,7,12,18,6,13,11,11,7,7,4,6,8,8,4,4,5,7,3,4,2,3,0,0
0,1,2,1,2,1,3,2,2,6,10,11,5,9,4,4,7,16,8,6,18,4,12,5,12,7,11,5,11,3,3,5,4,4,5,5,1,1,0,1
0,1,1,3,3,2,6,2,5,9,5,7,4,5,4,15,5,11,9,10,19,14,12,17,7,12,11,7,4,2,10,5,4,2,2,3,2,2,1,1
0,0,2,0,4,2,2,1,6,7,10,7,9,13,8,8,15,10,10,7,17,4,4,7,6,15,6,4,9,11,3,5,6,3,3,4,2,3,2,1
0,1,1,3,3,1,3,5,2,4,4,7,6,5,3,10,8,10,6,17,9,14,9,7,13,9,12,6,7,7,9,6,3,2,2,4,2,0,1,1
We want to:
- Load data into memory,
- Calculate the average value of inflammation per day across all patients, and
- Plot the results.
To do all that, we’ll have to learn a little bit about programming.
Loading Data
Let’s import the file called inflammation.csv into our R environment.
To import the file, first we need to tell our computer where the file is.
We do that by choosing a working directory, which is possible using
setwd("~/Desktop/FAIR-R/"). However, a more elegant way is creating an RStudio
project: File > New project > Existing Directory > Browse to the
folder you created during the setup.
.Rproj-fying your other workIf you have already worked with R before, but not with RStudio projects, repeat the above steps to upgrade a few of
Existing Directory. Note how double-clicking a resulting.Rprojfile launches RStudio right in the correct working directory.
Setting the working directory correctly is very important in R.
If it’s not, we’ll often get errors that files are not where we expect them to be.
That’s why the .Rproj files are more useful than using setwd() manually.
Now, we can load the downloaded data file into R:
(dat <- read.csv(file = "inflammation.csv", header = FALSE))
The expression read.csv(...) is a function call
that asks R to run the function read.csv and assign its output to a variable.
The ( parentheses ) around it instructs R to not only do the above, but also
show the function’s output in the console.
read.csv has two arguments: the name of the file we want to read, and whether the first line of the file contains names for the columns of data.
The filename needs to be a character string (or string for short), so we put it in quotes. Assigning the second argument, header, to be FALSE indicates that the data file does not have column headers. In case of our inflammation.csv example, R auto-generates column names in the sequence V1 (for “variable 1”), V2, and so on, until V30.
Other Options for Reading CSV Files
read.csvactually has many more arguments that you may find useful when importing your own data in the future. You can learn more about these options in this supplementary lesson.
Loading Data with Headers
What happens if you forget to put
header = FALSE? The default value isheader = TRUE, which you can check with?read.csvorhelp(read.csv). What do you expect will happen if you leave the default value? Before you run any code, think about what will happen to the first few rows of your data frame, and its overall size. Then run the following code and see if your expectations agree:read.csv(file = "inflammation.csv")
Reading Different Decimal Point Formats
Depending on the country you live in, your standard can use the dot or the comma as decimal mark. Also, different devices or software can generate data with different decimal points. Take a look at
?read.csvand write the code to load a file calledcommadec.txtthat has numeric values with commas as decimal mark, separated by semicolons.
A function will perform its given action on whatever value is passed to the argument(s).
For example, in this case if we provided the name of a different file to the argument file, read.csv would read that instead.
We’ll learn more about the details of functions and their arguments in the next lesson.
Since we included the whole line in parentheses, the console will display the full contents of the file inflammation.csv, which we read in assigned to the variable dat.
Try it out.
We can think of a variable as a container with a name, such as x, current_temperature, or subject_id that contains one or more values.
We can create a new variable and assign a value to it using <-.
Once a variable is created, we can use the variable name to refer to the value it was assigned. The variable name now acts as a tag.
To see the value of a variable, we can print it by typing the name of the variable and hitting Return (or Enter).
In general, R will print to the console any object returned by a function or operation
unless we assign it to a variable, or force-print the assignment’s output with
( parentheses around the whole epxression).
Variable Naming Conventions
Historically, R programmers have used a variety of conventions for naming variables. The
.character in R can be a valid part of a variable name; thus the above assignment could have easily beenweight.kg <- 57.5. This is often confusing to R newcomers who have programmed in languages where.has a more significant meaning. Today, most R programmers 1) start variable names with lower case letters, 2) separate words in variable names with underscores, and 3) use only lowercase letters, underscores, and numbers in variable names. The book R Packages includes a chapter on this and other style considerations.
When you assign a value to a variable, R only stores the value, not the calculation you used to create it. This is an important point if you’re used to the way a spreadsheet program automatically updates linked cells. Let’s look at an example.
For large data sets it is convenient to use the function head to display only the first few rows of data.
head(dat)
V1 V2 V3 V4 V5 V6 V7 V8 V9 V10 V11 V12 V13 V14 V15 V16 V17 V18 V19 V20
1 0 0 1 3 1 2 4 7 8 3 3 3 10 5 7 4 7 7 12 18
2 0 1 2 1 2 1 3 2 2 6 10 11 5 9 4 4 7 16 8 6
3 0 1 1 3 3 2 6 2 5 9 5 7 4 5 4 15 5 11 9 10
4 0 0 2 0 4 2 2 1 6 7 10 7 9 13 8 8 15 10 10 7
5 0 1 1 3 3 1 3 5 2 4 4 7 6 5 3 10 8 10 6 17
6 0 0 1 2 2 4 2 1 6 4 7 6 6 9 9 15 4 16 18 12
V21 V22 V23 V24 V25 V26 V27 V28 V29 V30 V31 V32 V33 V34 V35 V36 V37 V38
1 6 13 11 11 7 7 4 6 8 8 4 4 5 7 3 4 2 3
2 18 4 12 5 12 7 11 5 11 3 3 5 4 4 5 5 1 1
3 19 14 12 17 7 12 11 7 4 2 10 5 4 2 2 3 2 2
4 17 4 4 7 6 15 6 4 9 11 3 5 6 3 3 4 2 3
5 9 14 9 7 13 9 12 6 7 7 9 6 3 2 2 4 2 0
6 12 5 18 9 5 3 10 3 12 7 8 4 7 3 5 4 4 3
V39 V40
1 0 0
2 0 1
3 1 1
4 2 1
5 1 1
6 2 1
Manipulating Data
Now that our data are loaded into R, we can start doing things with them.
First, let’s ask what type of thing dat is:
class(dat)
[1] "data.frame"
The output tells us that is a data frame. Think of this structure as a spreadsheet in MS Excel that many of us are familiar with. Data frames are very useful for storing data and you will use them frequently when programming in R. A typical data frame of experimental data contains individual observations in rows and variables in columns.
What if we need the average inflammation of each patient, or the average for each day?
avg_patient_inflammation <- rowMeans(dat)
And to obtain the average inflammation of each day we will need to calculate the mean of all of the columns of the data frame.
avg_day_inflammation <- colMeans(dat)
Versatile Alternatives
The common row- or column-wise
mean()operations have the above shortcuts. But what if you need to perform other calculations? In base R, the family ofapplyfunctions will help you do that, seehelp(apply)or?apply. You can also learn about the modern approach with purrr::map().
Plotting
The mathematician Richard Hamming once said, “The purpose of computing is insight, not numbers,” and the best way to develop insight is often to visualize data. Visualization deserves an entire lecture (or course) of its own, but we can explore a few of R’s plotting features.
Let’s take a look at avg_day_inflammation using the function plot.
plot(avg_day_inflammation)
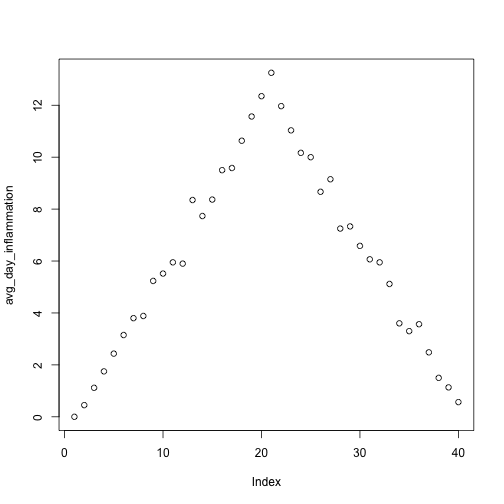
Above, we gave the function plot a vector of numbers corresponding to the average inflammation per day across all patients.
plot created a scatter plot where the y-axis is the average inflammation level and the x-axis is the order, or index, of the values in the vector, which in this case correspond to the 40 days of treatment.
The result is roughly a linear rise and fall, which is suspicious: based on other studies, we expect a sharper rise and slower fall.
So, either there’s a mistake in our calculations or something is wrong with our data.
In the next episode, we’ll create some functions to help us
Plotting Data
Create a plot showing the average inflammation for each patient across all days.
Key Points
Objects are created on demand whenever a value is assigned to them.
applying the mean calculation across rows or columns has shortcuts, but other statistics may need?applyor?purrr.Use
plotto create simple visualizations.