Pre-Configuring Advanced Searches and Retrieving the Results
Katrin Leinweber
2019-06-28
Source:vignettes/pre-configuring-advanced-searches-and-retrieving-the-results.Rmd
pre-configuring-advanced-searches-and-retrieving-the-results.RmdUnfortunately, the BacDive Web Service does not allow SQL-like queries for the content of specific fields within the strain’s datasets. If you find the functionality explained in BacDive-ing in too limited, please try the following, semi-automatic approach to using BacDiveR.
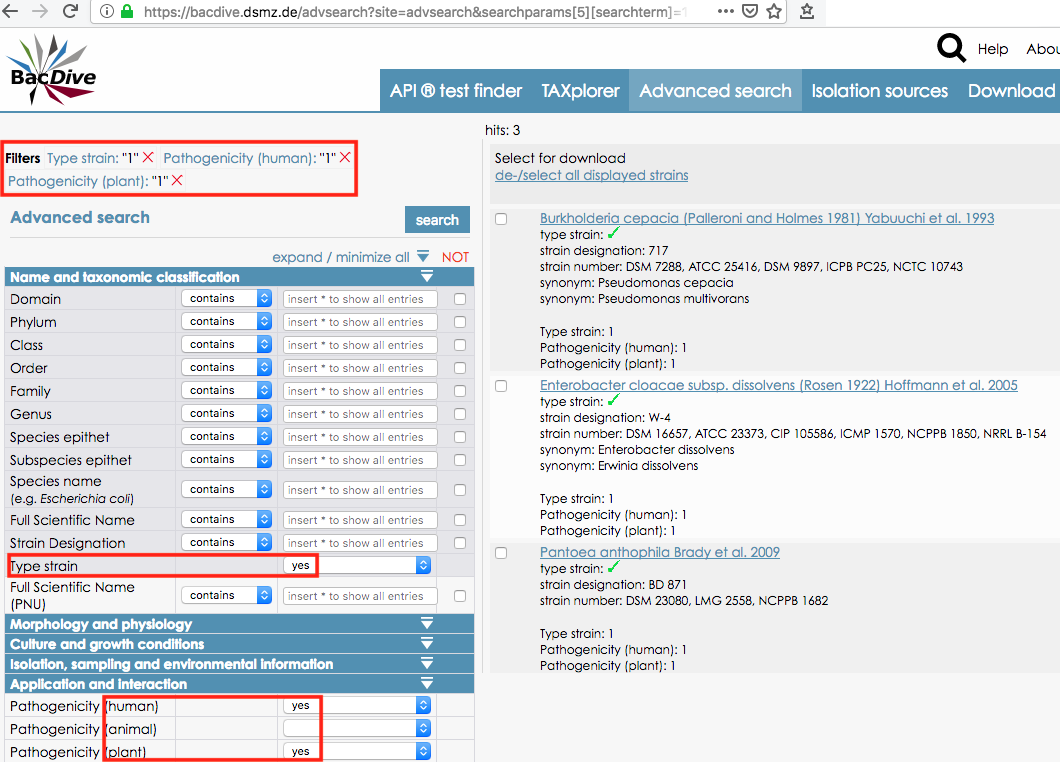
- Visit BacDive.DSMZ.de/AdvSearch and prepare your search in that web interface. It enables SQL-like searches against the ca. 135 of BacDive’s accessible data fields.
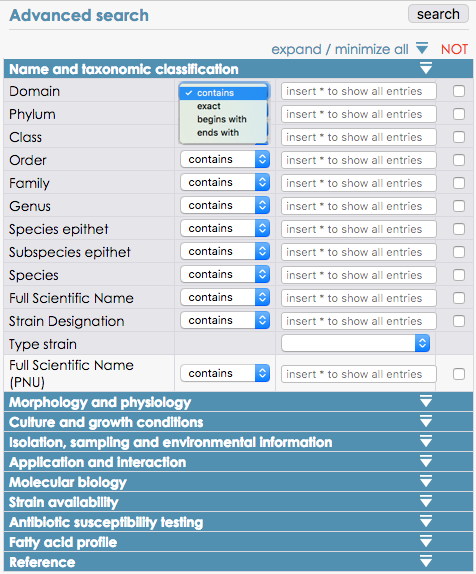
Overview of the possible fields to query and their parameters (contains, exact, begins/ends with)
- Run your advanced search (query). The example below searches for all strains that type a species pathogenic to both plant and human. Note the results list with the “hits” on the right, and the now much longer URL. It contains/encodes all the terms and parameters of your advanced search.
Copy the URL of the results page from your browser’s address bar. Alternatively, copy it from the “Download list of BacDive Ids” link to the top right of the “hits” list.
Paste the copied URL into a call to the
data <- bd_retrieve_by_search("…")function.Enjoy the list of downloaded datasets, just as you would after using
data <- bd_retrieve_data(searchTerm = ..., searchType = ...).
Mass-downloading datasets
bd_retrieve_data(searchTerm = …, searchType = "taxon") can be used to download all datasets for the genus or a specific species given in …. Broader searches are possible through the advanced search, for example for all Archaea:
Archaea_data <- bd_retrieve_by_search("https://bacdive.dsmz.de/advsearch?advsearch=search&site=advsearch&searchparams%5B70%5D%5Bcontenttype%5D=text&searchparams%5B70%5D%5Btypecontent%5D=contains&searchparams%5B70%5D%5Bsearchterm%5D=archaea")
Please note the messages about estimated download times for such large downloads.
Storing datasets offline
This is not a BacDiveR feature, but base R’s saveRDS() is particularly useful for offline-storage of lots of search results, because re-downloading them would take rather long. Continuing the Archaea example, the following code writes the dataset to a file, loads it again, and verifies the data integrity: